C All quantitative events for a single protein; quantitative ratios are provided numerically and also represented, in log space, by a slider. The publisher's final edited version of this article is available at J Proteome Res. The Qurate application itself simply manages the lists and arrays of data underlying the charts, which are all generated using the charting API in the platform. PepXML files containing database search results may also be written as well as read, allowing for filtration of database search results, normalization of quantitative ratios, etc also provided. This module is called an "ElutionCurveStrategy". Open in a separate window. Qurate charts for quantitative events. 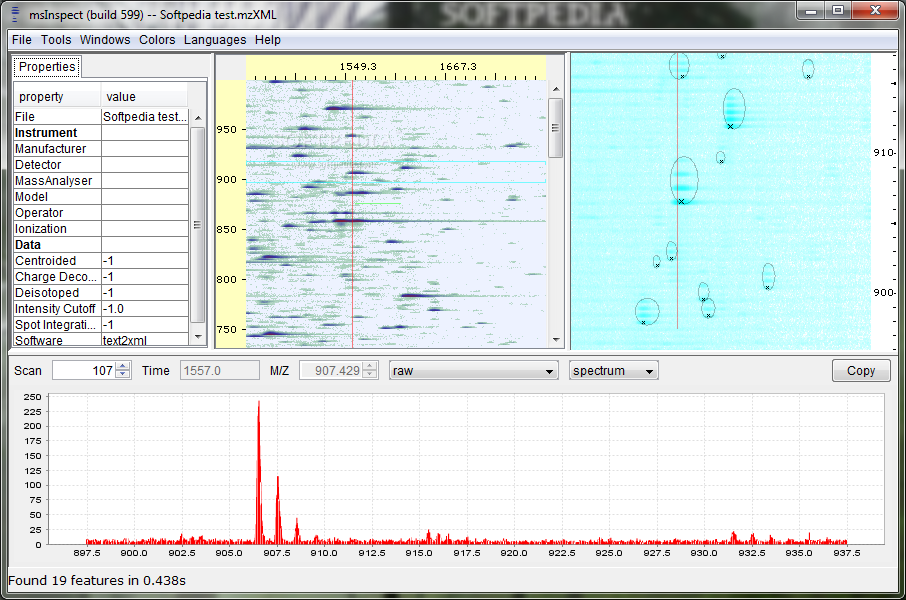
| Uploader: | Tolkree |
| Date Added: | 15 May 2015 |
| File Size: | 9.35 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 53029 |
| Price: | Free* [*Free Regsitration Required] |
The Qurate application msknspect simply manages the lists and arrays of data underlying the charts, which are all generated using the charting API in the platform. When this file is present, MRMer calculates and displays the light-to- heavy ratio of the elution AUCs areas under the elution curves.

Blue indicates high intensity. We used msInspect platform tools to remove all identified peptides with probability less than 0. It also provides a robust framework for creating applications that may be invoked from the command line or via a graphical interface, removing much of msinsprct software development burden from the researcher.

To mimic the process of msijspect differential candidates, we selected 50 quantitative events or groups of equivalent events, as described above from these 12 proteins with high light-to-heavy ratios; we also selected 50 quantitative events to serve as negative controls, from proteins with ratios close to parity 0. The msInspect platform is largely written in Java, with several statistical tasks performed in the R statistical language.
MRMer was developed in partnership with Dr. PepXML files containing database search results may also be written as well as read, allowing for filtration of database search results, normalization of quantitative ratios, etc also provided.
Our self-self quantitative dataset contained a total of high-confidence, quantified proteins with peptide identifications having probability greater than 0.
A software platform for rapidly creating computational tools for mass spectrometry-based proteomics
D A chart showing intensities near the quantitative event separately for each scan, with green and yellow lines indicating theoretical monoisotopic and other peaks; only the scan number in green is used by the quantitation algorithm for this event, meaning that the light peaks visible in the bottom scan were not used in ratio calculation.
B Summary information about all confidently identified, quantitated proteins. The purpose of this manuscript is to introduce the structure of the toolkit underlying the msInspect platform and to demonstrate its capabilities by creating a tool with practical utility in proteomics data analysis. Dashed lines indicate the first and last scans used by Q3, and X marks indicate peptide identification events. Use of the Platform to Build a Tool for Visual Curation of Quantitative Proteomics Experiments The msInspect software has been used to create several comprehensive applications 3 - 6but we discuss here the development of a tool to support visual inspection and curation of quantitative proteomics experiments.
It also provides a full-featured interface with the R statistical language. Specific use of msInspect Platform to develop Curate Command Module Framework The Qurate application is invoked either graphically or on the command line via a Command Module, using the Command Module framework described above.
A software platform for rapidly creating computational tools for mass spectrometry-based proteomics
These situations can all produce quantitative ratios that are extremely high or extremely low, erroneously causing the peptide in question to appear differentially abundant. Software Tools msInspect contains a number of tools that are not specific to proteomics data analysis but are intended to support rapid software development and the generation of general-use tools.
Qurate Self-Self Comparison Our self-self quantitative dataset contained a total of high-confidence, quantified proteins with peptide identifications having probability greater than 0. This structure also provides automatic documentation of the Command Module: Green and yellow lines indicate monoisotopic and other peaks.
Because the source code with documentation, a Developer's Guide describing platform tools in detail, and a support forum are available online at proteomics. Author information Copyright and License information Disclaimer.
We present this toolkit's capability to rapidly develop new computational tools by presenting an example application, Qurate, a graphical tool for manually curating isotopically msinspecf peptide quantitative events. This module is called an "ElutionCurveStrategy". The design and development of Qurate, including msinnspect revisions, consumed less than 5 days of total personnel time developer and data analystthanks to the extensive use of mwinspect tools already available in the platform.
Proteomics Data Modeling Qurate uses platform Java classes to model and manipulate protein identifications, peptide identifications and quantitative events, amino acid modifications, and LC-MS machine runs and spectra. The software, called Qurate, allows the user to examine labeled quantitation events, such as SILAC, ICAT or Acrylamide labeling 8visually, using several different informational charts, and to filter bad quantitation events out of the dataset.
msInspect - G6G Directory of Omics and Intelligent Software
The platform contains several File Management Utilities. False extreme ratios can stem from signal processing errors in the mzinspect ion scans, e. Peptide assignments were evaluated using PeptideProphet 9. Whenever one compares two complex samples in which most proteins do not change between conditions, one can expect that the extreme ratios will include many false positives due to errors in signal processing, including database matching and quantitation of ion signal intensities.
ProteinProphet may then be rerun in order to recalculate protein-level quantitative ratios using the remaining peptide quantitation events. Qurate was used to discover quantitative events that appeared, based on visual inspection, to be of low or questionable quality.

No comments:
Post a Comment